Prof. Dr. Ali Ahmad Naz Pflanzenzüchtung
- Telefon
- 0541969-5314
- a.naz@hs-osnabrueck.de
- Abteilung
- Fakultät Agrarwissenschaften und Landschaftsarchitektur
- Raum
- HP 0108
- Sprechzeiten
- Nach Vereinbarung
- Web
- Internetseite
- Beschreibung
- Fakultät Agrarwissenschaft und Landschaftsarchitektur
Vita
| 05/2015 | Habilitation as “venia legendi” in Plant Breeding and Genetics at the Department of Plant Breeding, Institute of Crop Sciences and Resource Conservation, University of Bonn, Germany Habilitation thesis: “Genetic mapping and positional cloning of genes controlling monogenic, oligogenic and polygenic traits in crop plants” |
| 09/2006 | PhD at the Department of Plant Breeding, Institute of Crop Sciences and Resource Conservation, University of Bonn |
| 2001 - 2002 | M.Sc (Honours) agriculture (horticulture) at the University of Agriculture Faisalabad, Pakistan |
| 1996 - 2000 | B.Sc (Honours) agriculture- specialization in horticulture at the University of Agriculture Faisalabad, Pakistan |
| Since 06/2021 | Professor of Plant Breeding at the University of Applied Sciences Osnabrueck Research and teaching focus: Plant breeding, genetic resource utility for sustainable crop production |
| 11/2009-05/2021 | Scientist (group leader) at the Department of Plant Breeding, Institute of Crop Sciences and Resource Conservation (INRES), University of Bonn, Germany Research focus: Biodiversity, biotic and abiotic stress tolerance in crops |
| 10/2006–09/2009 | Post-doctorial scientist at the Department of Plant Breeding and Genetics, Max Planck Institute for Plant Breeding Research Cologne Research focus: Breeding leaf and shoot traits in tomato |
| 06/2003–09/2006 | Research associate (PhD scholar), Institute of Crop Sciences and Resource Conservation, University of Bonn Research focus: Genetic analyses of disease resistances in bread wheat |
- Inivited guest editor for International Journal of Molecular Science (ISI ranked, 5 year IF: 4.1) for a special issue on Plant Breeding and Genetics
- Member reviewing board of BARD- The USA-Israel Binnation Agricultural Research & Development Fund, and National Science Centre Poland
- Acting as reviewer for dozens of high-ranked scientific journal in the area of plant biology, plant breeding and genetics and plant sciences
- Elected representative of the scientific staff in the executive board of Institute of Crop Sciences and Resource Conservation (INRES), University of Bonn
- Project manager of plant breeding for S1-laboratory; Gene techniques and biological safety ordinance- GenTSV
- Member of German Society for Plant Breeding (GPZ) and European Association for Research on Plant Breeding (EUCARPIA)
- Three months interdisciplinary course at the Center for Development Research (ZEF), University of Bonn. During this, I completed an interdisciplinary project with social anthropologist on intellectual property rights and patenting of genetic resources and written a term paper entitle “Who Own Those Genes”
- Secretary communication (Ex) Horticultural Society UAF-Pakistan
- Volunteer student leader for research extension to farming communities in Pakistan
Teaching & research concepts
Teaching concept
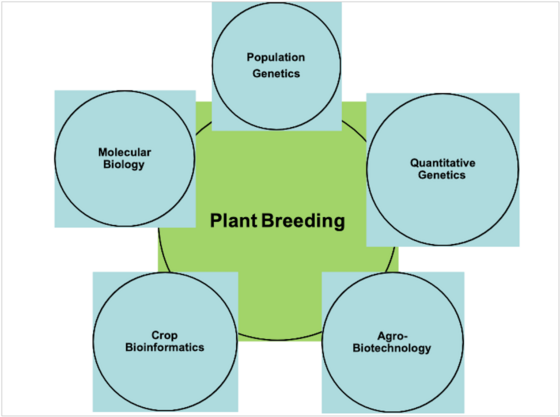
Plant breeding is a historical academic discipline which laid the foundations of modern agriculture. The principles and methods of classical breeding are still the nucleus of modern breeding science and industry. On the other hand, recent methodological advancements in genomics, biotechnology, molecular biology and bioinformatics have revolutionized the area of plant breeding and its linkages with the related disciplines. These developments have opened new inter-disciplinary areas of plant breeding with molecular genetics, population and quantitative genetics, biotechnology and bioinformatics. These development thus demand more a more dynamic curriculum of plant breeding encompasse its theoretical and application based linkages with associated academic disciplines. As plant breeding is an area of applied science, certainly there is a need to include more application based teaching curriculum combined with the possibility of research oriented teaching, practical, workshops, seminars and excursions.
Diagram showing synergies of plant breeding with the related academic disciplines.
Present research focus
Our research focuses on the interface of plant breeding and molecular genetics to explore, characterize and utilize vital traits in crop plants. The cultivated varieties are showing yield stagnancy and poor fitness against changing climate. In parallel, we are facing frequent drought and heat events, which are posing serious threats to crop production. Narrow diversity of cultivated gene pool seems a primary reason behind this void, thus demanding immediate attentions to advance research on natural diversity adapted to dry climates. For this, newly developed genomics and molecular tools offer efficient opportunities for a large-scale exploration and utilization of vital genetic resources in plant breeding. The recruitment of new diversity is essential to replenish the genetic potential of existing crop varieties for sustainable crop production in climate change scenario.
My present research is based on following strategy:
- Partitioning of breeding relevant trait variation into genetic and environmental variances using natural populations of crops plants for a precise and fast selection.
- Analyses of biotic and abiotic stress tolerances using genome-wide scans to identify breeding relevant quantitative trait loci, genes, epistasis and gene x environment interactions.
- Employment of molecular tools (CRISPR-Cas9 system) for innovative breeding goals and for molecular characterization of traits and underlying biological processes.
The primary objective of my research lies in the exploration and utilization of crop wild relatives for drought stress adaptation and disease resistance to improve yield sustainability in cereal crops. For drought stress adaptation, we investigated two fundamental processes; i) acquisition potential of water and ii) water use efficiency (conservation/utility) under drought stress. For acquisition potentials, root system variation is of great significance as morphological attributes facilitate the uptake of water and nutrients, favorable gas-exchange and carbon assimilation. In addition, root acts as primary sensor to detect abiotic stresses like drought and in response modulate stress adaptive mechanisms below ground as well as in aerial parts via root to shoot signal transduction. For this, we have identified novel variant for root system architecture from wild barley and incorporated favorable variation in cultivated varieties using genetic mapping and marker assisted selection approaches. For the water utility and conservation, we studied drought inducible synthesis of metabolite proline and its role in establishing physiological tolerance and stress recovery. Using molecular cloning and functional analyses, we have discovered a unique variation of pyrroline-5-carboxylate synthase (P5cs1) in barley diversity. P5cs1 is an enzyme-coding gene involved in the up-regulation of proline expression under drought stress conditions. The transfer of this P5cs1 exotic allele from wild barley to cultivated variety has shown potential in maintaining photosynthetic activity and grain yield under drought stress. In addition, the isogenic lines carrying exotic P5cs1 allele revealed potential for commercial synthesis of proline as agro-biotech and pharmaceutical product inside the crop plants. Further, we have employed genome-editing method CRISPR-Cas9 for the site-directed mutagenesis of P5cs1 gene and its driving transcription factors for in-depth molecular analysis and fine-tuning of water conservation and drought stress adaptation in barley. For disease resistance, we identified a number of novel alleles in wheat and barley diversity for the establishment of durable resistances against major fungal diseases.
Future research vision
I aim to extend my future research on the exploration and utilization of natural biodiversity for breeding relevant traits associated with yield and yield sustainability in cereal crops. Barley is considered as genomic model for cereals because of its self-pollinating and strict diploid behaviors. Its diversity reveals stunning morphological and physiological novelties for climatic adaptation because of its historical evolution across diverse environments. I believe genetic dissection of barley diversity provides an opportunity to identify and harness the useful genetic intelligence of un-adapted genepool for improving water use efficiency, abiotic and biotic stress tolerances in the cultivated varieties. This work in cereal model is essential to perform a targeted research in other related crops facing devastating losses due to drought and heat spells in recent years across the globe.
To accomplish this, I will employ methods of quantitative genetics and population genomics for in-depth evaluation of genes and genes by environment interactions. The power of quantitative genetics and genomics has increased manifold in the last decade by the advent of state of the art methods of genome analyses like next generation sequencing. By this, it became possible to identify genetic footprints of complex traits across genome with a great precision. In parallel, phenomics has emerged to look deep into whole plant level non-invasively for system-view of its performance under field conditions. New methods are available like transcriptomics, proteomics, metabolomics and ionomics termed as “molecular phenotyping” to quantify traits deeper at physiological levels. A high-resolution trait genetics and association of “omics” data showed potential to uncover genetic and molecular mechanisms behind the regulation of complex traits. Recently, site-directed mutagenesis using CRISPR/Cas9 system offered advantages for understanding the molecular and functional regulation of genes. Further, it is employed as an efficient tool in creating new variants and innovative breeding goals for their fast utility in plant breeding and related industries.
The specific objectives and steps of the proposed research are:
- Establishment of state of the art plant genetic resources in cereals for cutting edge research on biotic and abiotic stress tolerance in cereal and related species.
- Multi-scale phenotyping for high-resolution traits genetics using non-invasive phenotyping and “omics” methods across varying environmental (drought and heat stresses) conditions.
- Genetic dissection of trait variations using genome-wide scans approach to identify associated genes, genes epistasis and gene*environment interactions.
- Mining novel alleles from natural diversity of crops and their transfer to elite gene pool using marker assisted selection and speed breeding approaches.
- Site-directed genome-editing using CRISPR/Cas system for molecular and functional analysis of genes as well as for the creation of innovative breeding goals directly in cultivars.
- Understanding genetic and molecular mechanisms modulating water use efficiency and sustainability at morphological and physiological levels for establishing their synergistic link to enhance yield and sustainability.
Publications
- Shrestha A, Fendel A, Nguyen TH, Adebabay A, Kullik AS, Benndorf J, Leon J, Naz AA* (2022) Natural diversity uncovers P5CS1 regulation and its role in drought stress tolerance and yield sustainability in barley. Plant Cell Environ. (published on 21 September 2022) https://doi.org/10.1111/pce.14445
- Rasool F, Khan MR, Schneider M, Uzair M, Aqeel M, Ajmal W, Léon J, Naz AA* (2022) Transcriptome unveiled the gene expression patterns of root architecture in drought-tolerant and sensitive wheat genotypes. Plant Physiol. Biochem. 178:20-30
- Naz AA*, Bungartz A, Serfling A, Kamruzzaman M, Schneider M, Wulff BH, Pillen K, Ballvora A, Oerke EC. Ordon F, Léon J (2021) Lr21 diversity unveils footprints of wheat evolution and its new role in broad-spectrum leaf rust resistance. Plant Cell Environ. 44:3445–3458
- Chopra D, Hasan MS, Matera C, Chitambo O, Valerie-Mahlitz S, Naz AA, Janakowski S, Sobczak M, Mithöfer A, Kyndt T, Grundler F, Siddique S (2021). Parasitic worms redirect host metabolism via NADPH oxidase-mediated ROS to promote infection. New Phytol. 232:318–331
- Shrestha A, Cudjoe DK, Siddique S, Léon J, Naz AA* (2021). Abscisic acid binding transcription factors modulate proline biosynthesis and drought adaptation in Arabidopsis thaliana. J. Plant Physiol. 153414 (online). https://doi.org/10.1016/j.jplph.2021.153414
- Siddiqui MN, Léon J, Naz AA*, Ballvora A* (2020) Genetics and genomics of root system variation in adaptation to drought-stress in cereal crops. J. Exp. Bot. 72: 1007–1019, *Joint correspondence
- Muzammil S, Shrestha A, Dadshani S, Pillen K, Siddique S, Léon J, Naz AA* (2018) An ancestral allele of Pyrroline-5-carboxylate synthase1 promotes proline accumulation and drought adaptation in cultivated barley. Plant Physiol. 178: 771-782
- Bungartz A, Klaus M, Mathew B, Léon J, Naz AA* (2016) Development of first SNP derived cleaved amplified polymorphic sequence marker set and its successful utilization in the genetic analysis of seed color variation in barley. Genomics 107: 100-107
- Naz AA, Raman S, Martinez C, Sinha N, Schmitz G, Theres K (2013) Trifoliate encodes an R2R3 MYB transcription factor that modulates leaf and shoot architecture in tomato. Proc. Natl. Acad. Sci. USA 99: 1064-1069
- Naz AA, Kunert A, Volker L, Pillen K, Léon J (2008) AB-QTL analysis in winter wheat: II. Genetic analysis of seedling and field resistance against leaf rust in a wheat advanced backcross population. Theor. Appl. Genet. 116: 1095-1104
*Asterix represents applicant’s publication as corresponding author
Additional publications
- Schneider M, Vedder L, Oyiga B, Mathew B, Schoof, H, Léon J, Naz AA* (2022). Transcriptome profiling of barley and tomato shoot and root meristems unravels physiological variations underlying photoperiodic sensitivity. PLoS One (accepted PONE-D-22-07169R1)
- Boby M, Léon J, Sillanpää, MJ, Naz AA (2021). Importance of correcting genomic relationships in QTL mapping model with an advanced backcross population. G3: Genes| Genomes| Genetics jkab105. DOI: 10.1093/g3journal/jkab105
- Frimpong F, Anokye M, Windt CW, Naz AA, Frei M, Dusschoten DV, Fiorani F (2021) Proline-Mediated Drought Tolerance in the Barley (Hordeum vulgare L.) Isogenic Line Is Associated with Lateral Root Growth at the Early Seedling Stage. Plants 14:10(10):2177, doi: 10.3390/plants10102177
- Frimpong F, Windt CW, van Dussschoten D, Naz AA, Frei M, Fiorani F (2021) A wild allele of Pyrroline-5-carboxylate synthase1 leads to proline accumulation in spikes and leaves of barley contributing to improved performance under reduced water availability. Front. Plant Sci. 12, 633448, https://doi.org/10.3389/fpls.2021.633448
- Rustamova S, Shrestha A, Naz AA, Huseynova I (2021) Expression profiling of DREB1 and evaluation of vegetation indices in contrasting wheat genotypes exposed to drought stress. Plant Gene (online) https://doi.org/10.1016/j.plgene.2020.100266
- Begum H, Alam MS, Feng Y, Koua P, Ashrafuzzaman M, Shrestha A, Kamruzzaman M, Dadshani S, Ballvora A, Naz AA*, Frei M* (2020) Genetic dissection of bread wheat diversity and identification of adaptive loci in response to elevated tropospheric ozone. Plant Cell Environ. 43: 2650-2665, *Joint correspondence
- Oyiga BC, Palczak J, Wojciechowski T, Lynch JP, Naz AA, Léon J, Ballvora A (2020). Genetic components of root architecture and anatomy adjustments to water-deficit stress in spring Barley. Plant Cell Environ. 43: 692-711
- Reinert S, Osthoff A, Léon J, Naz AA* (2019) Population genetics revealed a new locus that underwent positive selection in barley. Int. J. Mol. Sci. 8:20(1)
- Sendek A, Karakoç C, Wagg C, Domínguez-Begines J, do Couto GM, van der Heijden MGA, Naz AA, Lochner A, Chatzinotas A, Klotz S, Gómez-Aparicio L, Eisenhauer N (2019) Drought modulates interactions between arbuscular mycorrhizal fungal diversity and barley genotype diversity. Sci. Rep. 9: 9650
- Naz AA*, Dadshani S, Ballvora A, Léon J (2019) Genetic analysis and transfer of favorable exotic QTL alleles for grain yield across D genome using two advanced backcross wheat populations. Front. Plant Sci. 10: 711
- Ashraf A, Rehman O, Muzammil S, Léon J, Naz AA*, Rasool F, Ali GM, Zafar Y, Khan MR (2019) Evolution of Deeper Rooting 1-like homoeologs in wheat entails the C-terminus mutations as well as gain and loss of auxin response elements. PLoS One 14(4): e0214145.
- Anwer MA, Anjam MS, Shah SJ, Hasan MS, Naz AA, Grundler F, Siddique S (2018) Genome-wide association study uncovers a novel QTL allele of AtS40-3 that affects the sex ratio of cyst nematodes in Arabidopsis. J. Exp. Bot. 69: 1805-1814
- Bedawy I, Dehne H-W, Léon J, Naz AA* (2018) Mining the global diversity of barley for Fusarium resistance using leaf and spike inoculations. Euphytica 214: 18
- Naz AA, Reinert S, Bostanci C, Seperi B, Léon J, Böttger C, Südekum KH, Frei M (2017) Mining the global diversity for bioenergy traits of barley straw: genome-wide association study under varying plant water status. Glob. Change Biol. Bioenergy 9: 1356-1369
- Khan MR, Khan I, Ibrar Z, Léon J, Naz AA* (2017) Drought-responsive genes expressed predominantly in root tissues are enriched with homotypic cis-regulatory clusters in promoters of major cereal crops. The Crop J. 5: 193-206
- Sayed MA, Hamada A, Léon J, Naz AA* (2017) Genetic mapping reveals novel exotic QTL alleles for seminal root architecture in barley advanced backcross double haploid population. Euphytica 213: 1-16
- Reinert S, Kortz A, Léon J, Naz AA* (2016) Genome-wide association mapping in the global diversity set reveals new QTL controlling root system and related shoot variation in barley. Front. Plant Sci. 7: 1061
- Arifuzzaman Md, Günal S, Bungartz A, Muzammil S, Afsharyan N, Léon J, Naz AA* (2016) Genetic mapping reveals broader role of Vrn-H3 gene in root and shoot development beyond heading in barley. PLoS One 11(7): e0158718.
- Abebe T, Naz AA*, Léon J. (2015) Landscape genomics reveal signatures of local adaptation in barley (Hordeum vulgare L.). Front. Plant Sci. 6: 813
- Naz AA*, Klaus M, Pillen K, Léon J (2015) Genetic analysis and detection of new QTL alleles for Septoria tritici blotch resistance using two advanced backcross wheat populations. Plant Breed. 134: 514-519
- Naz AA (2015). Genetic mapping and positional cloning of genes controlling monogenic, oligogenic and polygenic traits in crop plants. Habilitation Monograph: University of Bonn.
- Mohammed NEM, Said AA, Naz AA, Bauer A, Mathew B, Reinders A, Léon J (2014) Association mapping for shoot traits related to drought tolerance in barley. Int. J. Agric. Innov. Res. 3: 68-79
- Naz AA*, Arifuzzaman Md, Muzammil S, Pillen K, Léon J (2014) Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley (Hordeum vulgare L.). BMC Genet. 15: 107
- Arifzumann Md, Sayed MA, Muzammil S, Schumann H, Pillen K, Naz AA*, Léon J (2014) Detection and validation of novel QTL for shoot and root traits in barley (Hordeum vulgare L.). Mol. Breed. 34: 1373-1387
- Sayed MA, Schumann H, Pillen K, Naz AA, Léon J (2012) AB-QTL analysis reveals new alleles associated to proline accumulation and leaf wilting under drought stress conditions in barley (Hordeum vulgare L.). BMC Genet. 13: 61
- Naz AA, Ehl A, Pillen K, Léon J (2012) Validation for root-related quantitative trait locus effects of wild origin in the cultivated background of barley (Hordeum vulgare L.). Plant Breed. 131: 392-398
- Naz AA, Kunert A, Kerstin F, Pillen K, Léon J (2012) Advanced backcross quantitative trait locus analysis in winter wheat: Dissection of stripe rust seedling resistance and identification of favorable exotic alleles originated from a primary hexaploid wheat (Triticum turgidum ssp. dicoccoides x Aegilops tauschii). Mol. Breed. 30: 1219-1229
- Naz AA, Rafique T, Jaskani MJ, Abbas H, Qasim M (2007) In vitro studies on micrografting technique in two cultivars of citrus to produce virus free plants. Pak. J. Bot. 39: 1773-1778
- Naz AA (2007) Comparative mapping of genes for plant disease resistance in wheat 'advanced backcross' populations by means of DNA markers. Shaker Verlag Aachen, Germany ISBN: ISBN-10: 3832261834
- Kunert A, Naz AA, Dedeck O, Pillen K, Léon J (2007) AB-QTL analysis in winter wheat: I. Detection of favourable exotic alleles for baking quality traits introgressed from synthetic hexaploid wheat (Triticum turgidum ssp. dicoccoides × T. tauschii). Theor. Appl. Genet. 15: 683-695
Published book chapter
-
Naz AA* and Jaskani MJ (2017). Genomics in Horticulture. In Horticulture Science and Technology. Faisalabad, Pakistan; University of Agriculture Faisalabad. http://onlinebooks.uaf.edu.pk
Manuscripts in peer-review process (pre-prints available)
-
Schneider M, Vedder L, Mathew B, Schoof H, Léon J. Naz AA*. A comparative transcriptome atlas of root and shoot development genes in crop models barley and tomato. Under review in Sci. Rep. (Manuscript ID SREP-20-00416)
- Main conference of plant breeding society (GPZ) Breeding plants for tomorrow’s world – challenges and solutions on 12 to 14 September at the University of Düsseldorf.
- Invited seminar in the “Summertagung GFPi Abteilung Pflanzeninnovation” am 28-29 Juni, 2022.
- Invited as keynote speaker at 13th International Barley Genetics Symposium – 2020, Riga, Letvia (called off due to corona).
- Scientific seminar on “A novel wild allele improves drought adaptation and yield sustainability in cultivated barley” in International Symposium (Digital Breeding) of the Society for Plant Breeding (GPZ) in cooperation with Saatgut Austria. February 11-13, 2020 | Tulln – Austria
- Invited lecture on „Genome-Editing - Ein neues Werkzeug in der Pflanzenzüchtung“ in the workshop „Neue Methoden in der Tier- und Pflanzenzucht” on 19.12.2018 by the cooperation of the University of Bonn, Research Network NRW-Agrar and Ministry for Climate Protection, Environment, Agriculture, Conservation and Consumer Protection of the State of North Rhine-Westphalia
- Scientific seminar in INRES Symposium (2018) “Mining novel alleles for sustainability in cereal crops” on 20.09.2018 in Bonn, Germany
- Project meeting of the Community for the Promotion of Plant Innovation e.V. (GFPi) (2017) „Genetische und molekulare Analyse von Braunrost Resistenz in Weizen“ on 02.02.2017 in Quedlinburg, Germany
- Research and academic exchange Workshop (DAAD) (2016) „Quantitative genetics promise new opportunities for crop yield and sustainability“ on 28.07.2016 at Lahore University of Management Sciences (LUMS), Lahore, Pakistan
- Invited presentation to Community for the Promotion of Plant Innovation e.V. (GFPi) Project meeting (2016) „Genetische und molekulare Analyse von Braunrost in Weizen“ on 20.04.2016 in Quedlinburg, Germany
- Presentation on the innovation day organized by Federal ministry of Agriculture and Nutrition (2016). “Identification of closely linked markers for leaf and stripe rusts and description of virulence and avirulence among free land pathogen populations” on 25-26.10.2016 in Bonn
- German-Pakistani research cooperation Workshop (2015) „Plant genetic resources and sustainable development in Pakistan“ on 26.10.2015 in University of Bonn
- Habilitations colloquium (2015) „Nutzung genetischer Ressourcen zur Verbesserung von Pflanzenpopulationen” on 13.05.2015 at Faculty of Agriculture, University of Bonn
- Professorial lecture (2015) „Green revolution versus blue revolution - a future perspective of genetic resources“ on 12.06.15 at the University of Bonn
- Seminar Chinese delegation, Department of Plant Breeding, University of Bonn (2015) „Host-pathogen interaction and sustainable agriculture“ on 16.07.2015 in Bonn
- International Conference on Enhanced Genepool Utilization- Capturing wild relative and landrace diversity for crop improvement (2014). „QTL detection for root-related traits in wild barley introgression lines under drought stress conditions“ on 16-20.06.2014 in Cambridge, United Kingdom
- Invited presentation to Community for the Promotion of Plant Innovation e.V. (GFPi) Public meeting of the Department of Oil and Protein Crops (2014) “Root system variation and drought stress tolerance” on 03.- 04.06.2014 Campus Klein-Altendorf, Uni-Bonn.
- Research and development Workshop by DAAD (2013) „Genome analysis and prospects of biotechnology in agriculture“ on 11.01.2013 in University of Agriculture Faisalabad, Pakistan
- CROP.SENSe.net Summer School, University of Bonn (2013) „Gene regulation in plant development: Methods of gene identification and characterization” on 24.05.13 in Bonn
- INRES colloquium, Institute of Crop Sciences and Resource Conservation (INRES) University of Bonn (2012) „A conserved mechanism of leaf and shoot development in tomato” on 29.06.2012 in Bonn
- Agriculture faculty colloquium, University of Bonn (2011) „The art of scientific writing“ on 20.07.2011 in Bonn
- Invited lecture, Department of Plant Breeding, University of Bonn (2009) „Gene isolation via map-based cloning in crop plants“ on 30.06.2009 in Bonn
- German Plant Pathology Foundation (DPG); Conference on disease management and resistance breeding (2005) „QTL analysis of disease resistance in two advanced backcross populations of wheat“ on 05-06.12.2005 in Fulda, Germany